Graphic abstract. credit: Comparative Biochemistry and Physiology Part D: Genomics and Proteomics (2022). DOI: 10.1016/j.cbd.2022.101026
Griffith’s new research uses biochemical blood profiles of sea turtles as a tool to monitor population health in the wild.
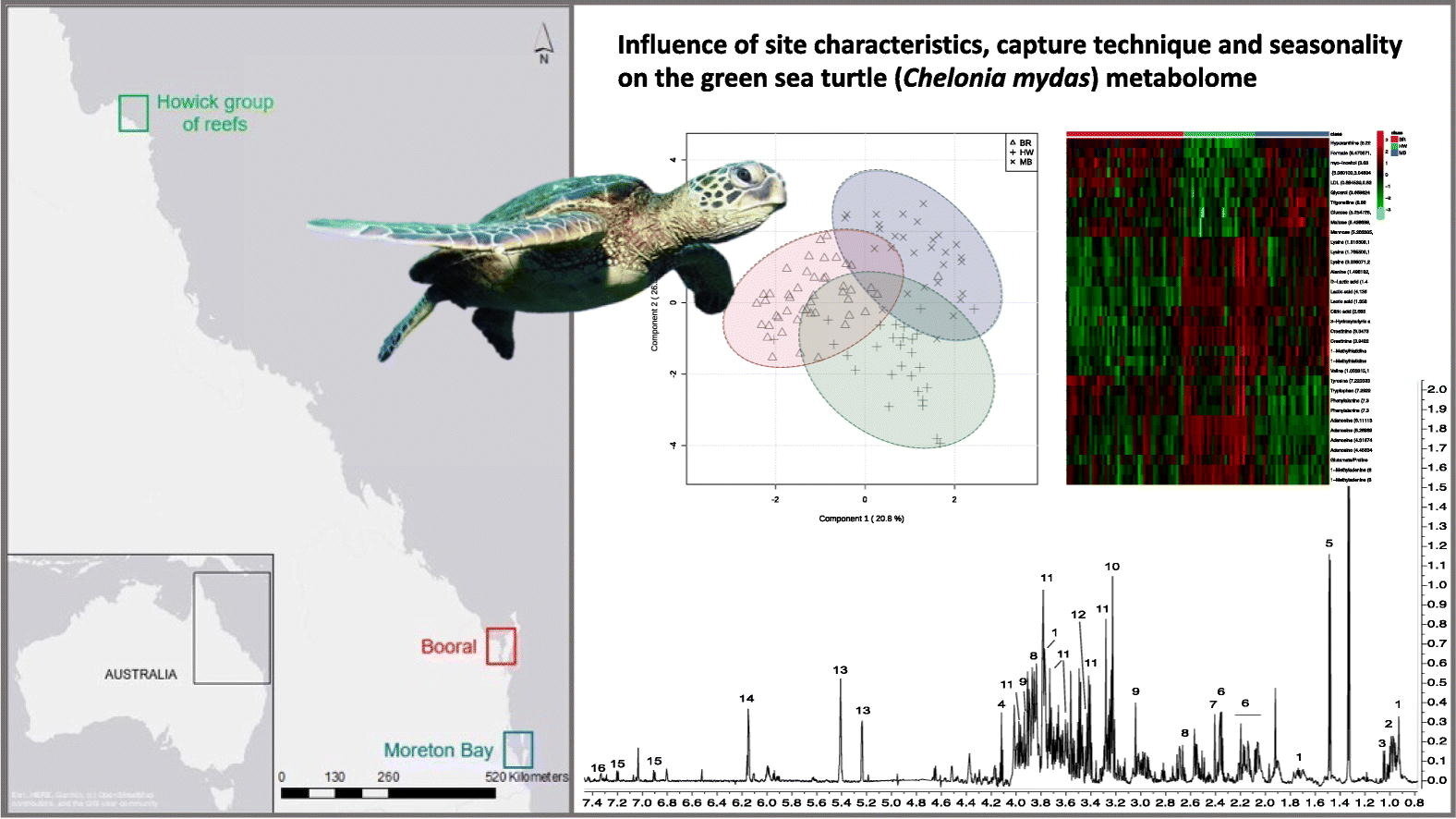
In the work published in Comparative Biochemistry and Physiology Part D: Genomics and Proteomicsresearchers used metabolomics, which measures the byproducts of physiological processes, to determine whether environmental conditions or how they were captured can affect their health.
“As an iconic but endangered species, there is considerable interest in the adaptation of advanced species analytical techniques to assess the health of wild sea turtle populations,” said Dr Steve Melvin, research fellow at the Australian Rivers Institute.
“Nuclear magnetic resonance spectroscopy is a powerful technique that can provide a metabolic fingerprint of physiological processes occurring in an animal. It provides a direct indication of the health of the organism and how external conditions affect the animal’s physiological response.”
“Being immortal, metabolomics provides an attractive method for comparing populations of endangered species, such as sea turtles, and for understanding how the environment in which they live affects their health. However, few studies have used this method to estimate wild sea turtle populations. .”
This is the first study of its kind to compare the biochemical profiles of both resting and active turtles from coastal and reef sites sampled over multiple seasons in South East Queensland.
“Our results show clear differences in the chemical fingerprints of turtles living in different locations, which can be attributed to different diets or forage quality and potentially different levels of exposure to stressors, such as chemical pollutants, between coastal and reef sites,” said Dr. Melvin.
“We also noticed clear markers of exercise in animals caught during activity using a method called ‘rodeo’, which were absent in turtles taken at rest.”
Turtles sampled from the same location over an 8-month period had only modest differences in metabolome over time, indicating the flexibility of the method and not prone to bias.
“When assessing the state of health of sea turtlesor other marine creatures in the wild, site characteristics such as animal quality’ source of foodthe amount of pollution and whether they are resting or active appear to have a much greater effect on turtle physiology than the effect of seasonal changes,” Dr Melvin said.
“Our study offers a real-world example of how a metabolomics technique, which provides a non-invasive snapshot of physiological health, can contribute to the monitoring and management of sea turtle populations and serve as an example for monitoring other marine megafauna. species over large areas and time scales in the wild.”
Stephen D. Melvin et al. Field monitoring of green sea turtles (Chelonia mydas): effects of site characteristics and capture technique on blood metabolome, Comparative Biochemistry and Physiology Part D: Genomics and Proteomics (2022). DOI: 10.1016/j.cbd.2022.101026
Provided
Griffith University
Citation: Biochemical Snapshot of Sea Turtle Health (2022, October 10) Retrieved October 10, 2022, from https://phys.org/news/2022-10-biochemical-snapshot-sea-turtle-health.html
This document is subject to copyright. Except in good faith for the purpose of private study or research, no part may be reproduced without written permission. The content is provided for informational purposes only.